Jupyter-JSC
Research and analysis of large amounts of data from scientific simulations, in-situ visualization and application control are convincing scenarios for interactive supercomputing. The open-source software Jupyter¹ is a tool that has already been successfully used in many scientific disciplines. With its open and flexible web-based design, Jupyter (or JupyterLab2) is ideal for combining a wide variety of workflows and programming methods under one interface. With the multi-user capability of Jupyter via JuypterHub3, it is suitable for use at supercomputing centers and thus combines the way HPC users work on the local laptop with the operation of a supercomputer. To ensure that each user has access to his data on the HPC systems UNICORE is used. The combination of the UNICORE tools XUUDB, UNICORE/X, Gateway and TSI allows JupyterHub to offer users to spawn Jupyter applications with their HPC accounts on all supported HPC systems. The integration of Unity-IdM4 into UNICORE enables Jupyter-JSC5 to delegate the login procedure to Unity-Idm. Therefore Jupyter-JSC is able to meet the desire for more interactivity in supercomputing and to open up new possibilities of high-performance computing. A simple, direct web access for starting and connecting with Jupyter or JupyterLab to the login or compute node was integrated at the Juelich Supercomputing Centre (JSC).
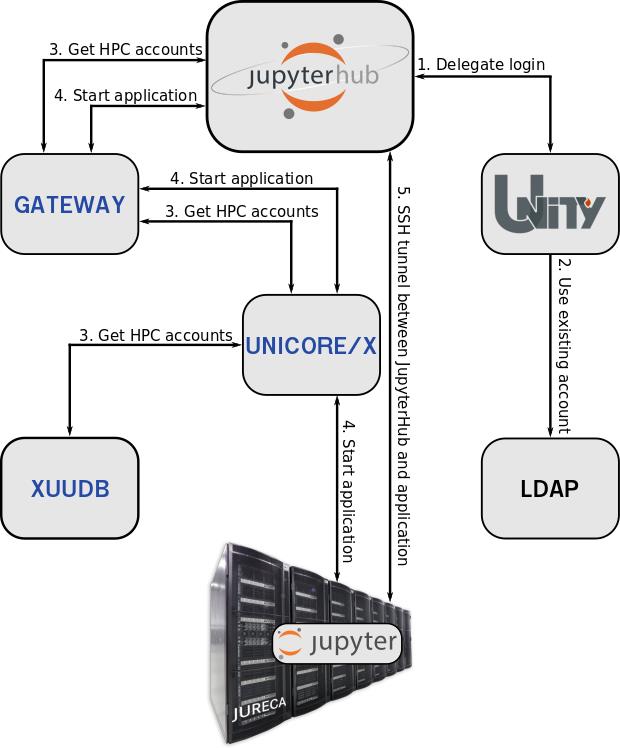
UNICORE enables Jupyter-JSC to start and manage jobs on the HPC systems at JSC. This ensures that every user has access to his data and can work with a Jupyter application on them. In addition, UNICORE offers reliable management of the Jupyter application. It provides access to all information output by the Jupyter application. Thanks to UNICORE, JupyterHub does not require direct access to the user’s HPC account. In practice Jupyter-JSC could not be operated without UNICORE.
References
[1] Jupyter: jupyter.org
[2] JupyterLab: github.com/jupyterlab/jupyterlab
[3] JupyterHub: github.com/jupyterhub/jupyterhub
[4] Unity project website: unity-idm.eu
[5] Jupyter-JSC: jupyter-jsc.fz-juelich.de
The human brain flagship
One particularly important and challenging application scenario is the European flagship Human Brain Project (HBP)1 . A very wide range of topics is targeted, ultimately aiming at a deeper understanding of the human brain, and attempting to leverage this deeper understanding for new technological advancements.
One particular area of work is the provisioning of a high-performance computing and data analysis platform. This comprises four major supercomputing sites in Juelich, Barcelona, Bologna and Lugano as well as cloud storage and other resources. An HTML5/JavaScript based web platform is being developed which will need to access these resources. Developers want to write custom applications for directly accessing services of the HPC Platform, such as job management or data transfer using REST API. The HBP uses a single-sign-on system based on OpenID Connect (OIDC)2 which enables users to use same account for accessing all services for which they have the required access permissions. Unity3 acts as a bridge to HBP OIDC infrastructure and provides a centralized authentication and delegation service for UNICORE infrastructures.
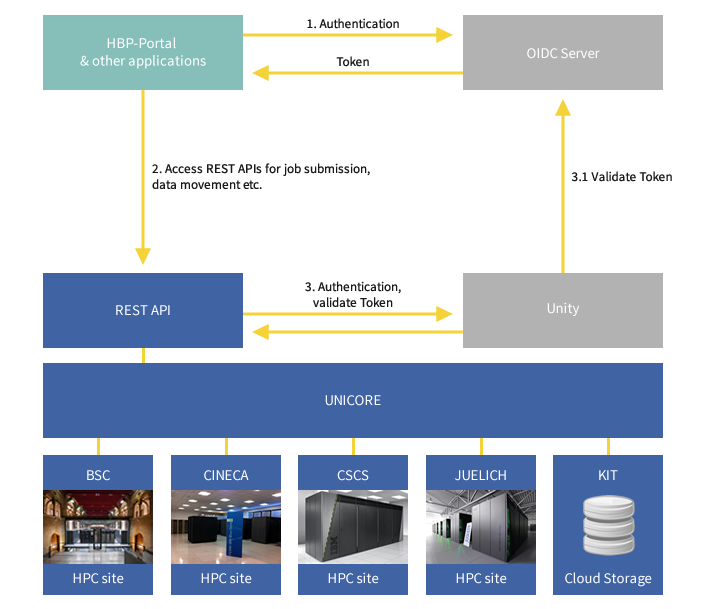
In Human Brain Project, UNICORE ensures seamless and secure access to the HPC and data resources from various European supercomputing sites. The following figure describes how UNICORE, the OIDC Server and Unity communicate in order to authenticate an HBP user. When a user wants to use HPC services via UNICORE, the identity is first verified by the OIDC server using the HBP username and password. The OIDC server returns an OIDC token in case of a successful authentication. This OIDC token is then used to access the UNICORE services. UNICORE passes this token to the UNITY server that validates the token by contacting the OIDC server. In case of a successful validation, the user can access the resources of the HPC Platform using the REST API.
References
[1] Human Brain Project: humanbrainproject.eu
[2] OpenID Connect: openid.net/connect
[3] Unity project website: unity-idm.eu
From single molecule to light emission
Nanomatch GmbH specializes in the simulation of organic thin-film morphologies, their electronic properties and interface structures such as bulk-heterojunctions. The company’s software allows materials discovery in a multi-scale approach building a morphology from a single molecule and calculates electronic properties such as electron or hole mobilities in a self-consistent density functional theory (DFT)-based approach.
Even though the multi-scale approach reduces the total computational effort , it still requires cluster resources to deliver results in adequate timescales. For a company the diverse availability of different cluster managers and job submission tools poses a large development task, which differs for each customer.
UNICORE is used as a unified and general grid API to allow the company’s software to run on the diverse landscape of clusters ranging from local test clusters to large supercomputing centres. Targeting only UNICORE allows the development of a single, tested code path and enables reproducibility by encapsulating the company’s products in workflows, which can be prepared by senior scientists and applied in large-scale material screening applications by junior-scientists.
The previous development efforts relied on the UNICORE Rich Client, but currently Nanomatch focuses on the REST API, which is used to develop a generalized workflow client including workflow inheritance.
One such workflow is the morphology generation workflow, which allows the generation of amorphous thin-film morphologies based on the atomic coordinates of a single molecule:
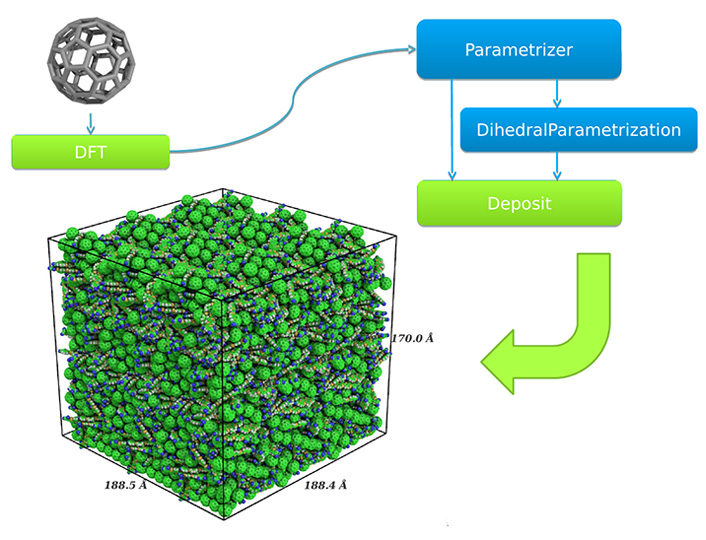
© Nanomatch GmbH
A morphology is generated only from the atomic coordinates of a molecule. After relaxing the molecule in a geometry relaxation DFT step, partial charges are calculated and possible dihedral angles are parametrized in a custom dihedral forcefield. The forcefield is then used in the linear-scaling deposition approach to generate a morphology shown in the lower left corner.
On the trail of brain-fibers
High-resolution three-dimensional polarized light imaging (3D-PLI) is an approach pursued by the Institute of Neuroscience and Medicine at the Forschungszentrum Jülich to create a detailed, three-dimensional map of nerve fibres of the human brain. Scientists strive to reach an understanding of the connectivity of the human brain as well as to study neurodegenerative diseases. In the 3D-PLI approach post-mortem brains are cut in thin slices (about 1500 slices, each 70 micron thick) and imaged with a microscope device using polarized light.
The images of brain slices are processed with a chain of tools for calibration, independent component analysis, enhanced analysis, stitching and segmentation. These tools have been integrated in a UNICORE workflow, exploiting many of the workflow system features, such as control structures and human interaction. Prior to the introduction of the UNICORE workflow system, the tools involved were run manually by their respective developers. Thus, once one step in the process was finished, the developer of the next tool in the chain would retrieve the data and run his tools on the output of the former. This manual approach led to delays in the entire process.
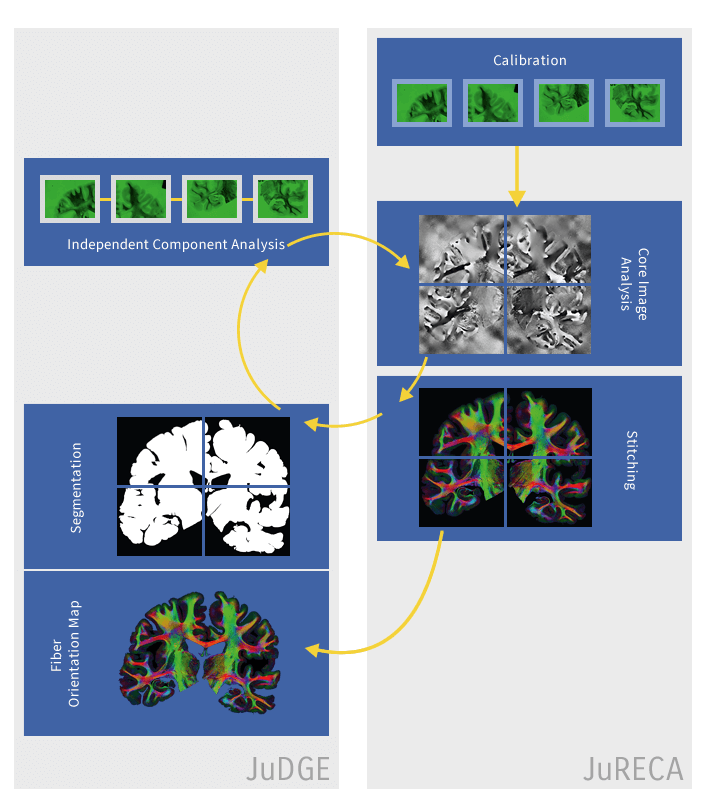
© Forschungszentrum Jülich
The UNICORE workflow system makes this approach almost fully automated and thus reduces the makespan of the entire workflow to hours rather than weeks. The results are easier to reproduce and the operation failures have been minimized, as fewer manual steps are involved. Only the automated approach will allow for the timely analysis of a large number of brain slices that are expected to be available in the near future.
